Electron Transfer and Transport in Nucleic
Acids
The ability to design and build artificial
supramolecular nanomachines that perform complex chemical
transformations with high efficiency and selectivity is an
important goal of modern chemistry. Toward this goal, this
project is exploring the use of synthetic nucleic acids
(NAs) to program the self-assembly of electroactive
structures that separate charge on the tens of nanometers
length scale. Single molecule conductance measurements are performed using a modified
version of the pioneering scanning tunneling microscope
break junction (STM-BJ) method in which the voltage applied
across the molecular junction has a triangular waveform. A
characteristic solvent response can be distinguished from
the molecular current response and the current responses can
be separated through a fit to a devised model circuit.
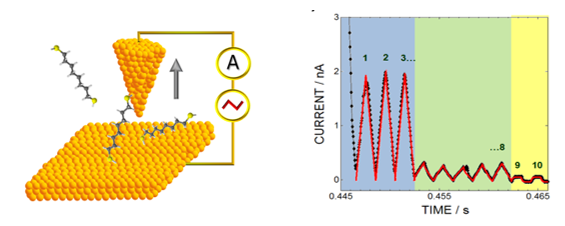
The left image
shows a schematic diagram illustrating the measurement of a
molecule’s conductance.
The right image shows a time trajectory for a single
octanedithiol molecule in a junction, which evolves from a
low resistance state (2x108 ohm) to a higher
resistance state (1.5x109 ohm) before the
junction breaks and gives only a capacitive response (yellow
region of time trajectory).
We are using this new
modulated bias protocol to determine the single molecule
conductance of nucleic acid duplexes as a function of backbone
and nucleobase sequence. PNA homoduplexes are found to exhibit a
larger conductance than DNA homoduplexes of the same sequence,
with DNA/PNA heteroduplexes having an intermediate conductance.
The findings support a conformationally-gated charge transfer
mechanism in which the increased flexibility of the PNA
homoduplexes facilitate electronic coupling.
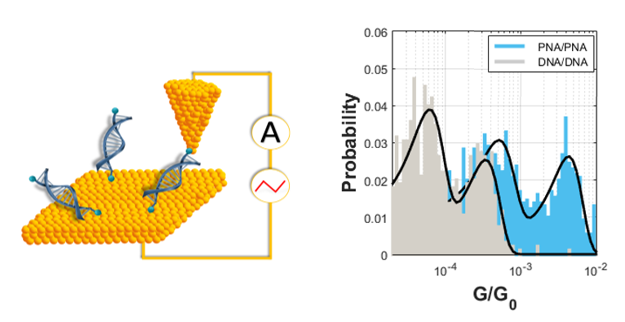
DNA/PNA
heteroduplexes will be the focus of future studies with the
long-term goal of studying redox-active moieties in precise 3D
arrangements on nucleic acid scaffolds.