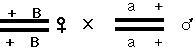
To finish off our FlyLab for this term, we will try to make complete linkage maps of the genes on chromosomes 1 (X), 2 and 3 of Drosophila. Bear in mind that we already have fairly complete map data for some of the genes on chromosome 3.
You have had enough experience by now to make your own decisions about these mapping experiments. You can do two-point crosses or three-point crosses as you see fit. You should make full corrections for double crossovers, of course. Note that in some cases you may not be able to include two genes in the same cross because of interactions between them and/or limitations in the FlyLab server. In such cases, do the best you can to infer the positions of each gene from the best experiments you can manage.
The one situation that may give you trouble is that of dominant alleles, some of which are known to be homozygous lethal. Here is an efficient strategy for two-point crosses involving recessive allele a and dominant alelle B. First cross:
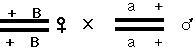
(If B is homozygous lethal, the female parent is heterozygous B/+; this doesn't matter, as all you want are the phenotypically B F1 females. Next, backcross phenotypically B F1 females to the male P:
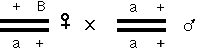
You can work it out that this is the equivalent of a testcross, so that recombinants are detected efficiently. Three-point crosses can be worked out using the same principle; I leave that to you.
When you have finished all your crosses, assemble your data into a complete recombination map. I have no objection if members of a group divide up the work, but it is a good idea to have each cross done independently by at least two people as a cross-check. We will pool data in class to construct complete maps for each chromosome.
Return to Honors Genetics Main Page