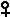 tra /+ x tra /tra
tra /+ x tra /tra
The following problems are to help you sharpen and deepen your understanding of sex determination, sex linkage and sex-linked inheritance. These questions will be discussed in the recitation section on Sept. 26. Problems are not to be handed in and will not be graded.
1. [Modified from Stansfield, Theory and Problems of Genetics ] An autosomal recessive gene (tra ), when homozygous, transforms a Drosophila female (XX) into a phenotypic male. All such "transformed" males are sterile. The gene has no effect in males (XY). You do the following cross:
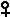 tra /+ x tra /tra
tra /+ x tra /tra
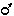
What is the expected sex ratio in the F1? If F1 males and females are chosen at random and mated, what is the expected sex ratio in the F2?
2. A rooster showing a mutant trait is crossed with a wild-type hen. You
observe that all female progeny have mutant phenotype and all male progeny have
wild-type phenotype. How would you describe the mode of inheritance of this
mutant trait? How does this result differ from what you would see in an
analogous cross (mutant
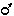 x wt
x wt
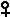 ) with Drosophila ?
) with Drosophila ?
3. [From Suzuki et al ., Introduction to Genetic Analysis ] Hypophosphatemia is caused by an X-linked dominant gene in humans. A man with hypophosphatemia marries a normal woman. What proportion of their sons will have hypophosphatemia?
4. The nematode Caenorhabditis elegans is normally a self-fertilizing hermaphrodite with XX sex chromosome composition. Rare males (XO) mate with hermaphrodites, such that the male's sperm compete with (but do not exclude) sperm from the hermaphrodite.
The unc-52 mutation produces a paralyzed phenotype when homozygous. You cross homozygous unc-52 hermaphrodites with wild-type males and observe the following classes of progeny:
5. The following observations were made in my own lab in September 1993. See if
you reach the same conclusion I did.
I had three strains of C. elegans in the lab:
Strain Genotype Linkage Group
EH16 lacZ X
PD55 lacZ V
CB224 dpy-11(e224) V
The dpy-11 mutation is recessive (phenotype Dpy when homozygous, non-Dpy when heterozygous). The "lacZ" genes are foreign genes ("transgenes") introduced from E. coli ; both EH16 and PD55 are homozygous for lacZ (albeit on different chromosomes), but there is no comparable gene in CB224. The lacZ gene product can be detected by an enzyme assay. The assay is sensitive enough so that hemizygotes (lacZ/0) can be detected with confidence.
I wanted to make a strain that was homozygous for both dpy-11(e224) and a lacZ gene. I therefore did the following cross:
EH16
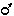 x CB224
x CB224

and picked individual non-Dpy F1 hermaphrodites to a new plate. The self-progeny of these F1 were about 75% non-Dpy and 25% Dpy. When tested for lacZ, none of the Dpy F2 had any lacZ activity. Some of the non-Dpy F2 did show lacZ activity, but I did not count them.
Return to Honors Genetics Main Page