|
Protein phosphorylation is estimated to affect 30% of the proteome and is
a major regulatory mechanism that controls many basic cellular processes.
Until recently, our biochemical understanding of protein phosphorylation
on a global scale has been extremely limited; only one half of the yeast
kinases have known in vivo substrates and for less than 160
phosphoproteins is the phosphorylating kinase known. Using proteome chip
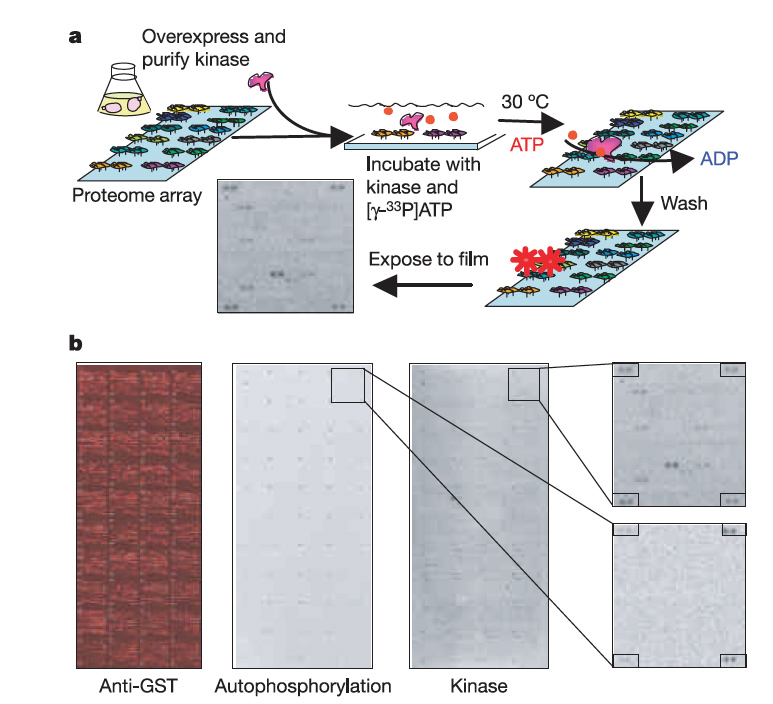
technology, we determined the in vitro substrates recognized by the
majority of yeast protein kinases. 4,192 phosphorylation events
involving 1,325 different proteins were identified; these substrates
represent a broad spectrum of different biochemical functions and cellular
roles. Distinct sets of substrates were recognized by each protein kinase,
including closely related kinases of the protein kinase A family and four
CDKs that vary only in their cyclin subunits. While many substrates reside
in the same cellular compartment or belong to the same functional category
as their phosphorylating kinase, many others do not, suggesting new roles
for a number of kinases. Furthermore, integration of the
phosphorylation results with protein-protein interaction and transcription
factor binding data revealed a number of novel regulatory modules. Our
phosphorylation results have been assembled into a first generation
phosphorylation map for yeast. Since many yeast proteins and pathways are
conserved, these results provide insights into the mechanisms and roles of
protein phosphorylation in many eukaryotes.
|
|
1Department of Molecular Biophysics & Biochemistry and
2Molecular, Cellular & Developmental Biology. Yale University.
New Haven, CT 06511, USA
3Invitrogen Corporation, 1600 Faraday Avenue, Carlsbad,
California 92008, USA
4Department of Medical Genetics and Microbiology, University of
Toronto, 1 Kings College Circle, Toronto, Ontario M55 1A8, Canada
5Banting & Best Department of Medical Research, University of
Toronto, Rm. 4285, Medical Sciences Bldg., 1 King's College Circle,
Toronto, Ontario M55 1A8, Canada
6Department of Molecular Genetics and Biochemistry, University
of Pittsburgh School of Medicine Pittsburgh, PA 15261, USA
7Division of Gene Regulation and Expression, School of Life
Sciences, University of Dundee, Dundee DD1 5EH, United Kingdom
8Department of Pathology, Yale University School of Medicine,
P.O. Box 208023 310 Cedar St. BML 342, New Haven, CT 06520-8023, USA
9Department of Biology, California Institute of Technology,
Pasadena, CA 91125, USA
10Department of Microbiology and Molecular Medicine, CMU,
University of Geneva,1211 Geneva, Switzerland
|