
Department of Molecular Genetics and Biochemistry
University of Pittsburgh School of Medicine
Pittsburgh, Pennsylvania 15261
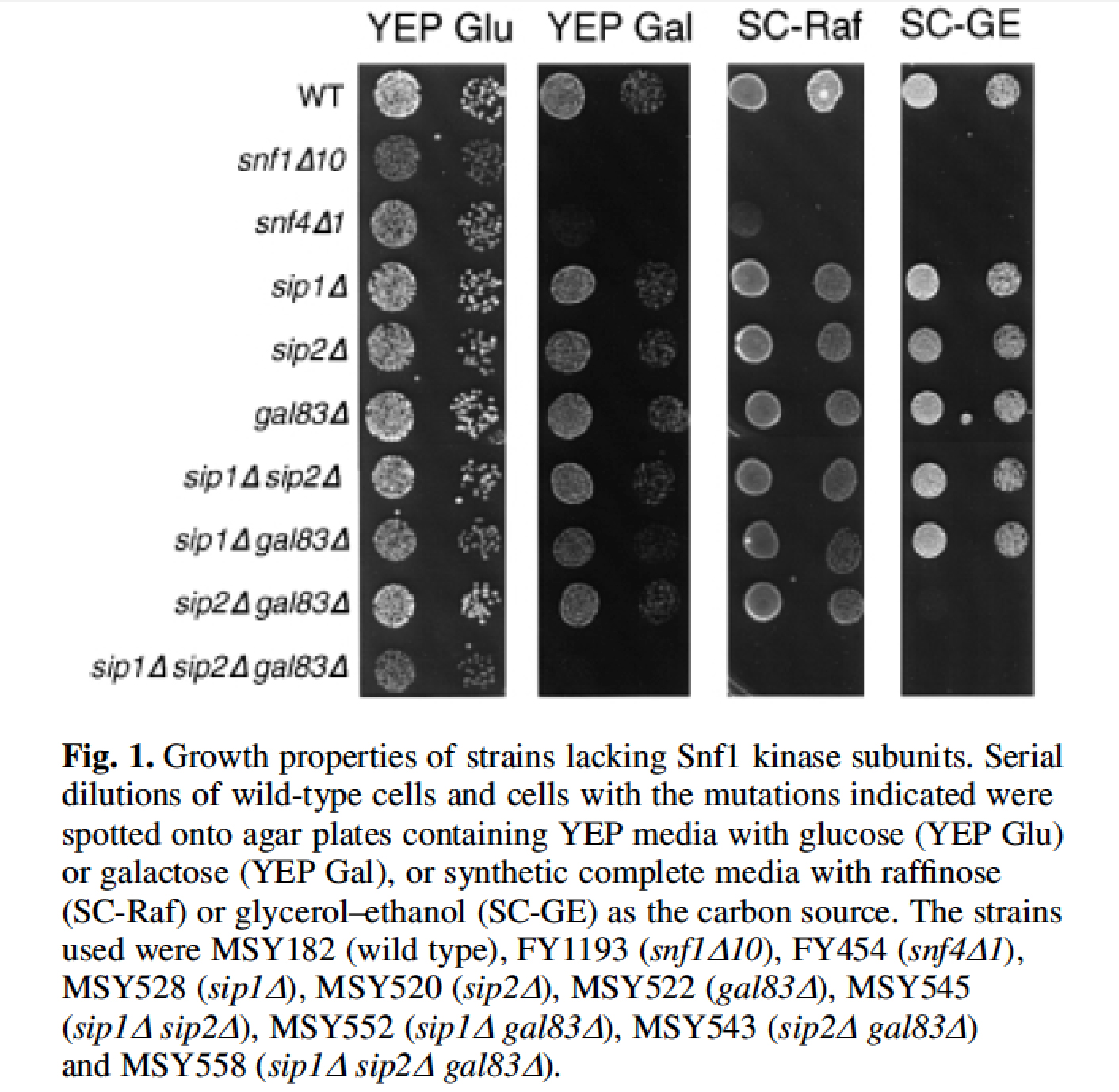
| The Snf1 kinase and its mammalian homologue, the AMP-activated protein kinase, are heterotrimeric enzymes composed of a catalytic alpha subunit, a regulatory gamma subunit and a beta subunit that mediates heterotrimer formation. Saccharomyces cerevisiae encodes three beta subunit genes, SIP1, SIP2 and GAL83. Earlier studies suggested that these subunits may not be required for Snf1 kinase function. We show here that complete and precise deletion of all three beta subunit genes inactivates the Snf1 kinase. The sip1 sip2 gal83 strain is unable to derepress invertase, grows poorly on alternative carbon sources and fails to direct the phosphorylation of the Mig1 and Sip4 proteins in vivo. The SIP1 sip2 gal83 strain manifests a subset of Snf phenotypes (Raf+, Gly-) compared to the snf1 strain (Raf-, Gly-), suggesting that individual beta subunits direct the Snf1 kinase to a subset of its targets in vivo. Indeed, deletion of individual of beta subunit genes causes distinct differences in the induction and phosphorylation of Sip4, strongly suggesting that the beta subunits play an important role in substrate definition. |
