Methodology and Fundamentals
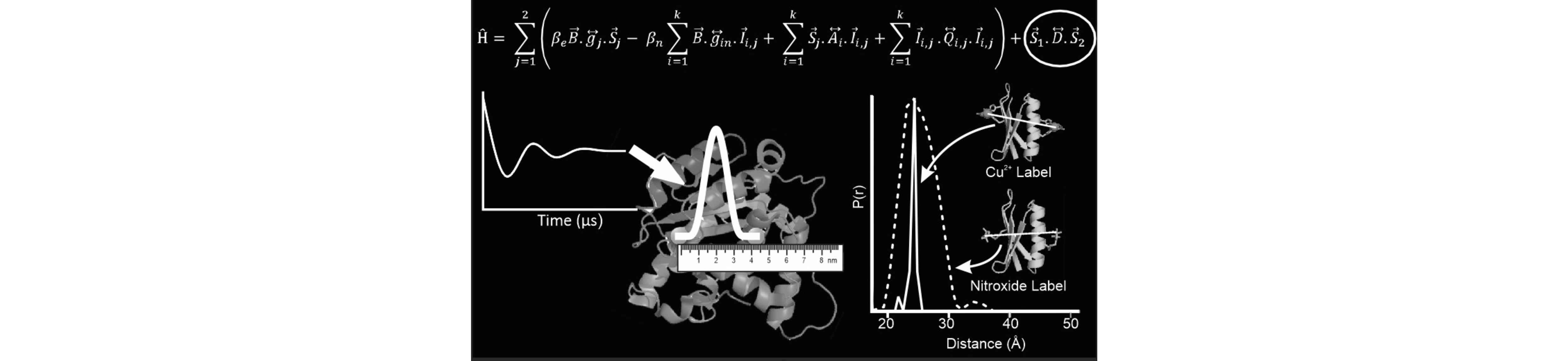
Electron Spin Resonance (ESR) has emerged as an important method for the measurement of structure-function relationships in proteins and nucleic acids. The basis for the use of ESR has resulted from the development of site directed spin labeling (SDSL). In SDSL of proteins a native amino acid is replaced with a cysteine by site directed mutagenesis. Thiol reactive methanethiolsulfonate (MTSSL), a common nitroxide spin label, is then chemically attached to the cysteine.
In this arena, our group pioneers the development of ESR methods to measure the dipolar interaction between two electron spins. In early work we recognized the potential of such methodology to measure large nanometer range distances in macromolecules. More recently, we have developed Cu(II)-ion based distance rulers that can measure nanometer range distance distributions in biomolecules. We have established a thorough understanding of double quantum and double resonance experiments for both Cu(II)-Cu(II) and Cu(II)-MTSSL interspin distance measurements. We have created analysis procedures to interpret experimental data in terms of distances. We have designed approaches where complications from electron-nuclear interactions are obviated in Cu(II) based distance measurements. In early experiments we also demonstrated the potential of Cu(II)-MTSSL labeling schemes for the measurement of distances at physiological temperatures. These ESR distance methods have proven attractive given that they are applicable to large molecular weight proteins, membrane proteins, large protein-complexes, and proteins exhibiting local/global protein conformational dynamics or high intrinsic disorder. Furthermore, in protein complexes, one can measure a sparse set of long range distances (a few nm) in order to measure the 3D assembly of different subdomains of a protein or a complex, and/or the amplitude of a structural change associated with function. The long-range distance constraints by ESR have also been combined with X-ray crystallography and NMR spectroscopy to elucidate the structure of large complexes.
Site-Directed Cu(II) Labeling
Electron spin resonance in combination with site-directed spin labeling has developed into a widely used technique to provide a multifaceted view of protein structure and dynamics. Our group has pioneered the use of paramagnetic metal ions like Cu(II) as an important spin label for both proteins and DNA.
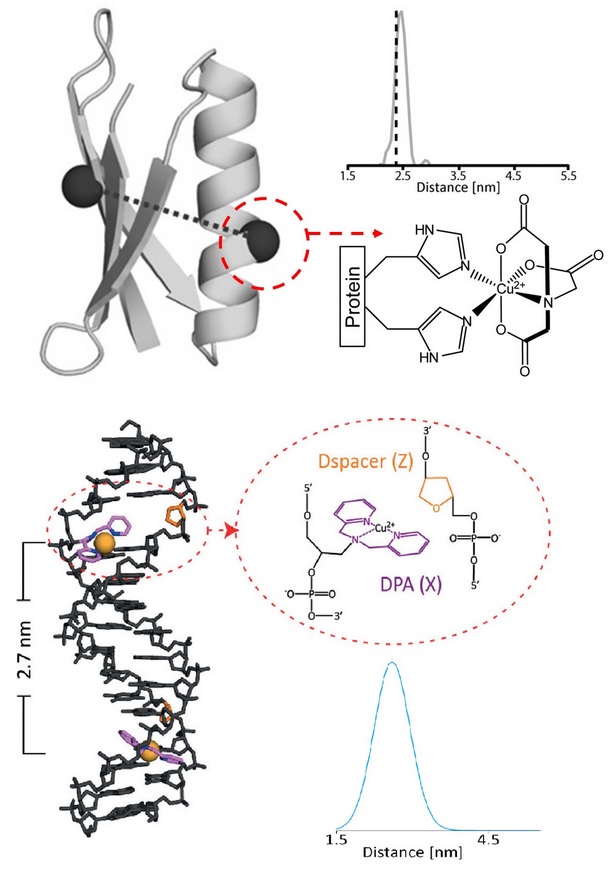
We have developed methodology to bind Cu(II) to specific sites on a protein in order to measure structural constraints that better reflect backbone structure and flexibility, compared to commonly used spin labels. In this method, the ESR reporter [i.e. a Cu(II) ion] is site specifically attached to the protein using a double histidine (dHis) mutation. For an α-helical site, an i and i+4 placement of the histidines accounts for the turn of the helix and produces a known Cu(II)-binding environment. For β-sheets, an i and i+2 residue placement creates a dHis site by placement of both histidines on the solvent-exposed face of the sheet. Non-specific binding of Cu(II) is prevented by introduction of the Cu(II)-ion as a complex with iminodiacetic acid (IDA) or nitrilotriacetic acid (NTA). Notably, the labeling is achieved by exploiting naturally occurring amino acids and does not require post-expression synthetic modification, other than addition of the Cu(II)-IDA or Cu(II)-NTA complex. The Cu(II) position is significantly restricted by coordination to protein side-chains and, therefore, the resultant distances are remarkably precise, with a distance distribution width that is as much as five times narrower when compared to the nitroxide spin label. This development constitutes a decisive improvement in labeling methodology that is not only simple, but also capable of providing unambiguous, highly relevant protein structural constraints.
For DNA, our group has developed an approach to site-specifically label DNA with Cu(II) for ESR distance measurements. In this approach, a 2,2’- dipicolylamine phosphoramidite is incorporated into any DNA oligonucleotide during initial DNA synthesis, which serves to chelate the ESR reporter [i.e. a Cu(II) ion]. The method only requires the simple post-synthetic addition of Cu(II) without the need for further chemical modification. Our approach places the ESR probe at the interior of the DNA helix, as opposed to ~1-1.5 nm outside in the helical perimeter as seen in current nitroxide-based labeling techniques. Therefore, the measured distance using current spin labels is as much as 1-2 nm greater than the biologically relevant distance. Detailed modeling is, therefore, necessary in order to extract distances that relate to DNA structure, often leading to ambiguities. In comparison, distance measurements based on our approach directly report on backbone distance and are maximally sensitive to DNA structure and flexibility. Importantly, our spin-labeling approach is structure and/or nucleotide independent.