

Next: The action potential
Up: Modeling synapses
Previous: Modeling synapses
In attempting to
model the behavior of whole neurons, detailed microscopy and staining
are required to produce a full three-dimensional picture of the
cell. This picture is then broken down into a series of cylinders with
varying diameters and lengths. These are then used to create a
mathematical and computational description of the cell. In order to
write equations for the compartments, we must determine the axial and
the transmembrane resistances as well as the membrane capacitance.
For a given cylindrical compartment,
the membrane resistance is related to the area of the compartment as
is the capacitance. Given a specific resistivity in Ohm-cm2, RM the
total resistance is just this value divided by the area, Rm=RM/A
whare A is the area. Similarly, if there are active channels with
conductances in millisiemens per square centimeter, the total
conductance is the specific conductance times the area. (Note that the
resistance is divided by the area and the conductance multiplied -
more area means more conductance and less resistance.) The
capacitance for the compartment is also proportional to the area.
Figure 1:
Two different cylinders coupled by the average axial
resistance
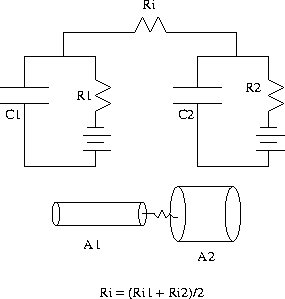 |
However, when we connect two compartments together, how do we define
the axial resistance between them if they have different lengths
and/or diameters? Obviously, if the two cylinders are identical, then
you can use either of the axial resistances as they are both the
same. If they are not the same, then there are several different
possibilities:
- If both are cylinders, average the two resistances and use the
average.
- If one is a cylinder and the other is a sphere, use the
cylindrical value
- Order the compartments and use the resistance corresponding to
the lower or higher number in the ordering
We will always use the first 2 algorithms. GENESIS lets you choose
which algorithm to use. I am not sure how NEURON resolves this.
EXAMPLE
Suppose that the two compartments have dimensions (length X diameter)
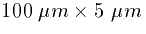 and
and 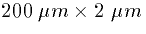 respectively. Then
respectively. Then
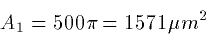
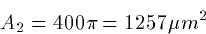
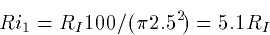
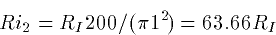
Ri = 34.4 RI
Thus, the two equations are
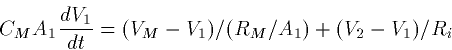
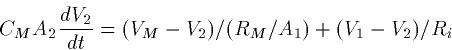
Dividing these by the area, we get
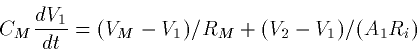
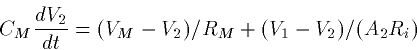
In other words the two equations for the voltages look identical
except for the coupling strength between them. (If the specific
resistances and potentials and channel densities were different for
the two cells, the noncoupling coefficients would also be different.)
The key point is that the effect of a big compartment on a little
compartment is asymmetric. In this example, since compartment 1 is
larger than compartment 2, the influence of compartment 2 on
compartment 1 is less than that of compartment 1 on compartment 2
since the former coupling is divided by a bigger number. Often in
modeling a 2 compartment system with compartment 1 bigger than
compartment 2, it is convenient to write the
couplings as:
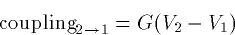
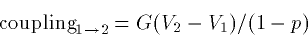
where G is a fixed conductance and p=1-A2/A1 is an asymmetry
parameter. p=0 if the compartments have the same area. As p gets
closer to 1, the ratio of the small to the big goes to zero. Big
dendrites have strong effects on little somas. This is why when the
soma spikes, there is little propagation back up the dendrite unless
the dendrites themselves have active channels.
Figure 2:
A 4 compartment model representing a pyramidal cell
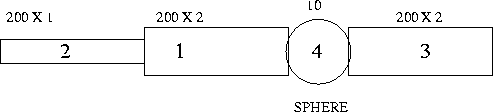 |
Homework
Write the equations for a compartmental model of a neuron with 4
compartments that are arranged as follows with dimensions given in
the figure (length and diameter in microns).
Compartments 1 and 2 are apical dendrites with
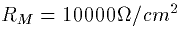 ,
, 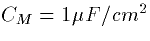 ,
, 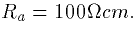 Compartment 3 is
a basal dendrite
with the same properties and compartment 4
is the spherical
soma that has a transient sodium conductance and delayed
rectifier as well as a leak. Assume that it has a transient sodium channel with a density
of 100 mS/cm2 a persistent potassium channel with density of
80mS/cm2, a leak conductance .1mS/cm2 and
VNa=50mV,VK=-100mV,Vl=-67mV. (Note that if one uses
millisiemens and microfarads as the dimensions, then the times will
be in milliseconds which is convenient.) (Also note that a leak
conductance of 0.1mS/cm2 corresponds to a membrane resistance of
Compartment 3 is
a basal dendrite
with the same properties and compartment 4
is the spherical
soma that has a transient sodium conductance and delayed
rectifier as well as a leak. Assume that it has a transient sodium channel with a density
of 100 mS/cm2 a persistent potassium channel with density of
80mS/cm2, a leak conductance .1mS/cm2 and
VNa=50mV,VK=-100mV,Vl=-67mV. (Note that if one uses
millisiemens and microfarads as the dimensions, then the times will
be in milliseconds which is convenient.) (Also note that a leak
conductance of 0.1mS/cm2 corresponds to a membrane resistance of
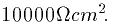 )
You do not have to simulate this (yet); I am mainly interested in seeing
that you understand how to put the compartments together with the
dynamics. The resting potential is -67mV.
)
You do not have to simulate this (yet); I am mainly interested in seeing
that you understand how to put the compartments together with the
dynamics. The resting potential is -67mV.


Next: The action potential
Up: Modeling synapses
Previous: Modeling synapses
G. Bard Ermentrout
2/12/1998

![]()
![]()
![]()
![]()
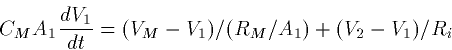
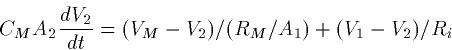
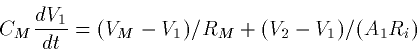
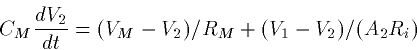
![]()
![]()